```{r echo = FALSE}
#| message: false
#| warning: false
pacman::p_load(tidyverse, readxl, knitr, kableExtra, performance, parameters,
latex2exp, see, patchwork, mfp, multcomp, emmeans, janitor, effectsize,
broom, ggmosaic, tinytable, ggrepel, glue, ggtext, ggdag,
conflicted)
conflicts_prefer(dplyr::select)
conflicts_prefer(dplyr::filter)
cb_pal <- c("#000000", "#E69F00", "#56B4E9",
"#009E73", "#F0E442", "#F5C710",
"#0072B2", "#D55E00", "#CC79A7")
cbbPalette <- cb_pal
theme_marginal <- function() {
theme_minimal() +
theme(panel.grid.minor = element_blank(),
plot.background = element_rect(fill = "white", color = NA),
plot.title = element_text(size = 16, face = "bold"),
plot.subtitle = element_text(size = 12, face = "italic"),
plot.caption = element_text(face = "italic"),
axis.title = element_text(face = "bold"),
axis.text = element_text(size = 12),
strip.text = element_text(face = "bold"),
strip.background = element_rect(fill = "grey80", color = NA))
}
```
# Baustelle {#sec-construction}
*Letzte Änderung am `r format(fs::file_info("construction-zone.qmd")$modification_time, '%d. %B %Y um %H:%M:%S')`*
{fig-align="center" width="100%"}
::: {.callout-caution appearance="simple"}
## Stand des Kapitels: Konstruktion (seit 07.2025)
Dieses Kapitel wird in den nächsten Wochen geschrieben. Ich plane zum Beginn des WiSe 2025/26 eine neue Version des Kapitels erstellt zu haben. Während das Kapitel entsteht, funktioniert so manches dann nicht so wie es soll.
:::
::: {.callout-caution appearance="minimal"}
## Dieses Kapitel ist archiviert
{fig-align="center" width="100%"}
Ich benötige die Thematik aktuell nicht in meiner Lehre oder der statistischen Beratung. Mir ist es als Nachschlagewerk aber immer noch wichtig zu behalten. Archivierte Kapitel werden nicht von mir weiter gepflegt oder ergänzt. Auftretende Fehler werden aber natürlich beseitigt, wenn die Fehler mir auffallen oder gemeldet werden.
:::
## Visualisierung von Verteilungen
- `rnorm()`: Erzeugt Zufallszahlen aus einer [Normalverteilung](https://en.wikipedia.org/wiki/Normal_distribution).
- `runif()`: Erzeugt Zufallszahlen aus einer [Gleichverteilung](https://en.wikipedia.org/wiki/Continuous_uniform_distribution).
- `rbinom()`: Generiert Zufallszahlen aus einer [Binomialverteilung](https://en.wikipedia.org/wiki/Binomial_distribution).
- `rpois()`: Erzeugt Zufallszahlen aus einer [Poisson-Verteilung](https://en.wikipedia.org/wiki/Poisson_distribution).
- `rgamma()`: Generiert Zufallszahlen aus einer [Gamma-Verteilung](https://en.wikipedia.org/wiki/Gamma_distribution).
- `rexp()`: Erzeugt Zufallszahlen aus einer [Exponentialverteilung](https://en.wikipedia.org/wiki/Exponential_distribution).
- `rt()`: Generiert Zufallszahlen aus einer [Student's t-Verteilung](https://en.wikipedia.org/wiki/Student%27s_t-distribution).
- `rchisq()`: Erzeugt Zufallszahlen aus einer [Chi-Quadrat-Verteilung](https://en.wikipedia.org/wiki/Chi-squared_distribution).
- `rbeta()`: Erzeugt Zufallszahlen aus einer [Beta-Verteilung](https://en.wikipedia.org/wiki/Beta_distribution).
- `rf()`: Erzeugt Zufallszahlen aus einer [F-Verteilung](https://en.wikipedia.org/wiki/F-distribution).
- `rlogis()`: Erzeugt Zufallszahlen aus einer [logistischen Verteilung](https://en.wikipedia.org/wiki/Logistic_distribution).
- `rweibull()`: Erzeugt Zufallszahlen aus einer [Weibull-Verteilung](https://en.wikipedia.org/wiki/Weibull_distribution).
```{r}
#| message: false
#| echo: false
#| warning: false
#| fig-align: center
#| fig-height: 4
#| fig-width: 7
#| fig-cap: "foo. *[Zum Vergrößern anklicken]*"
#| label: fig-gen-data-weibull
ggplot() +
theme_marginal() +
geom_vline(xintercept = c(0)) +
geom_hline(yintercept = c(0)) +
stat_function(fun = dweibull, linewidth = 1, args = list(scale = 1, shape = 1.5),
xlim = c(0, 8.25), aes(color = "1"), show.legend = FALSE) +
stat_function(fun = dweibull, geom = "area", args = list(scale = 1, shape = 1.5),
alpha = 0.25, xlim = c(0, 8.25), aes(fill = "1")) +
ylim(0, 2.5) + xlim(0, 3) +
scale_color_okabeito() +
scale_fill_okabeito()
```
```{r}
#| message: false
#| echo: false
#| warning: false
#| fig-align: center
#| fig-height: 4
#| fig-width: 7
#| fig-cap: "foo. *[Zum Vergrößern anklicken]*"
#| label: fig-gen-data-beta
df <- tibble(dist = c("a", "b", "c"), beta1 = c(1,3,5), beta2 = c(3,3,3))
df |>
group_by(dist) |>
reframe(x = seq(0, 1, 0.01), y = dbeta(x, beta1, beta2)) |>
ggplot(aes(x, y, color = dist)) +
theme_marginal() +
geom_line() +
scale_color_okabeito(labels = c(expression(beta[1]*","~beta[2]), expression(beta[1]),expression(beta[1])))
```
## Daten generieren
```{r}
library(ggeffects)
```
```{r}
set.seed(1234)
x <- rnorm(200, 5, 1)
z <- rnorm(200)
# quadratic relationship
# y <- 5 + 2 * x + x^2 + 4 * z + rnorm(200)
y <- 5 + 2 * (x-5) + (x-5)^2 + 4 * z + rnorm(200)
y <- x^2 - 8*x + 4*z +20 ## das war Gemini KI
d <- data.frame(x = x, y, z)
m <- lm(y ~ x + z, data = d)
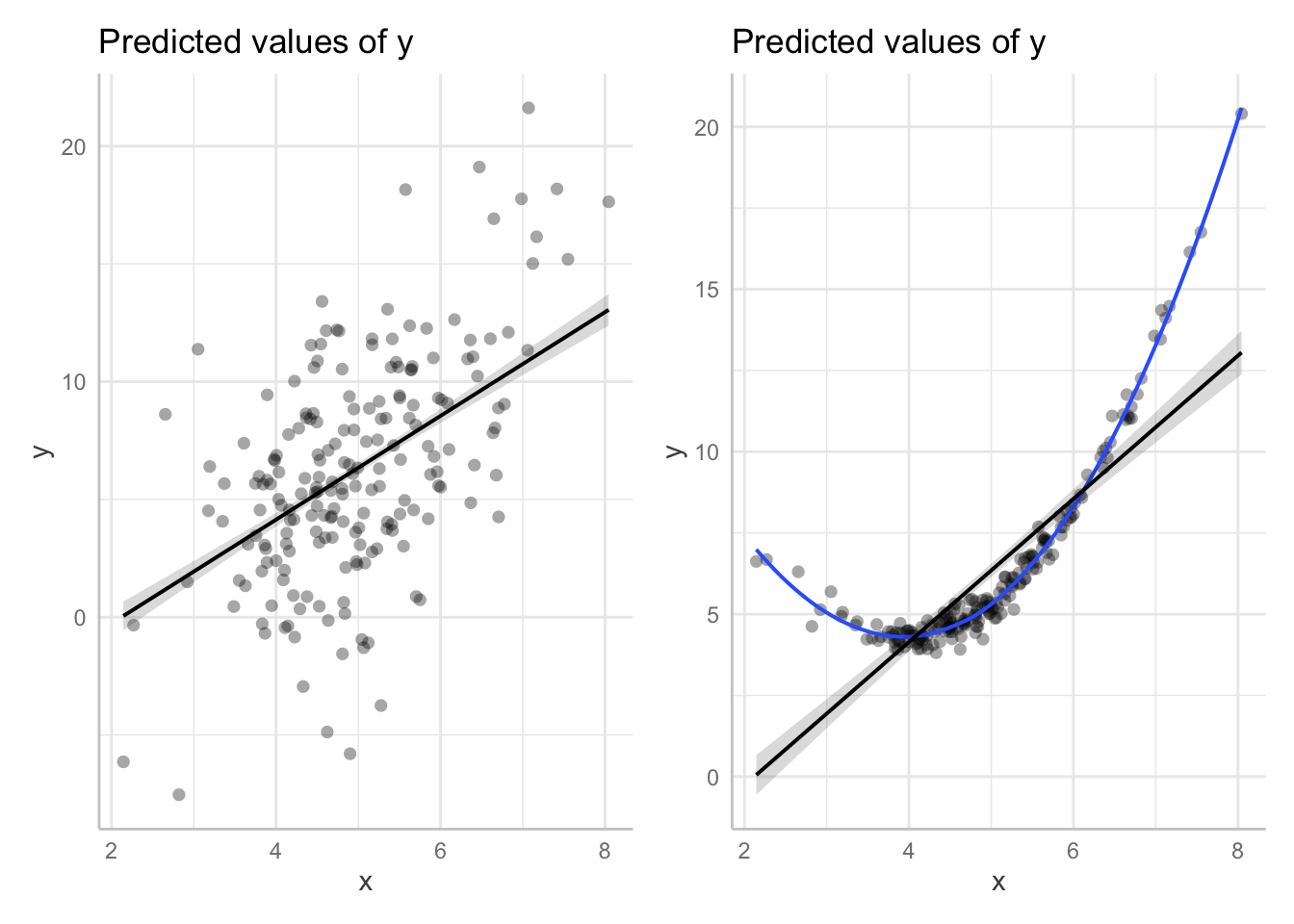
pr <- predict_response(m, "x [all]")
p1 <- plot(pr, show_data = TRUE)
p2 <- plot(pr, show_residuals = TRUE, show_residuals_line = TRUE)
p1 + p2
```
```{r}
x <- seq(-3, 3, length.out = 200) + rnorm(200) + 5
z <- rnorm(200)
y <- 5 + 2 * x + x^2 + x^3 + 4 * z + rnorm(200)
y <- 5 + 2 * (x-5) - (x-5)^2 + (x-5)^3 + 4 * z + rnorm(200)
y <- x^3 - 16*x^2 + 87*x + 4*z - 155
d <- data.frame(x = x, y, z)
m <- lm(y ~ x + z, data = d)
pr <- predict_response(m, "x [all]")
p1 <- plot(pr, show_data = TRUE)
p2 <- plot(pr, show_residuals = TRUE, show_residuals_line = TRUE)
p1 + p2
```
[Introduction: Adding Partial Residuals to Adjusted Predictions Plots](https://strengejacke.github.io/ggeffects/articles/introduction_partial_residuals.html)
### ... mit `{modelbased}`
[R Paket `{modelbased}`](https://easystats.github.io/modelbased/)
## Maximum Liklihood
- [In Statistics, Probability is not Likelihood.](https://www.youtube.com/watch?v=pYxNSUDSFH4) - [Maximum Likelihood, clearly explained!!!](https://www.youtube.com/watch?v=XepXtl9YKwc)
- [Likelihood!](https://kevintshoemaker.github.io/NRES-746/LECTURE4.html)
- [Maximum likelihood estimation from scratch](https://alemorales.info/post/mle-nonlinear/)
## Konfidenzintervall vs. Prädiktionsintervall
Ein Konfidenzintervall gibt den Wertebereich für einen gesuchten Parameter der Grundgesamtheit mit einer bestimmten Wahrscheinlichkeit an. Ein Prognoseintervall gibt den Wertebereich für einen individuellen, zukünftig zu beobachtenden Wert mit einer bestimmten Wahrscheinlichkeit an.
Konfidenzintervall
: Ein Konfidenzintervall gibt den Wertebereich für einen gesuchten Parameter der Grundgesamtheit mit einer bestimmten Wahrscheinlichkeit an.
Prädiktionsintervall
: Ein Prädiktionsintervall (auch Vorhersageintervall oder Prognoseintervall) gibt den Wertebereich für einen individuellen, zukünftig zu beobachtenden Wert mit einer bestimmten Wahrscheinlichkeit an.
[Quantile Regression Forests for Prediction Intervals](https://www.bryanshalloway.com/2021/04/21/quantile-regression-forests-for-prediction-intervals/)
[The difference between prediction intervals and confidence intervals](https://robjhyndman.com/hyndsight/intervals/)
[P-values for prediction intervals](https://robjhyndman.com/hyndsight/forecasting-pvalues.html) machen keinen Sinn
## Concordance Correlation Coefficient (CCC)
*Kann auch in technische Gleichheit mit rein*
```{r}
nirs_wide_tbl <- read_excel("data/nirs_qs_data.xlsx") |>
clean_names()
nirs_long_tbl <- nirs_wide_tbl |>
pivot_longer(cols = jd_ts:last_col(),
values_to = "values",
names_to = c("method", "type"),
names_sep = "_") |>
mutate(gulleart = as_factor(gulleart),
method = as_factor(method),
type = as_factor(type))
```
[Technical note: Validation and comparison of 2 commercially available activity loggers](https://www.sciencedirect.com/science/article/pii/S0022030218302418)
[User's guide to correlation coefficients](https://pmc.ncbi.nlm.nih.gov/articles/PMC6107969/)
[Concordance correlation coefficient calculation in R](https://medium.com/@amorimfranchi/concordance-correlation-coefficient-calculation-in-r-98d74ae5f0fc)
[How does Polychoric Correlation Work? (aka Ordinal-to-Ordinal correlation)](https://www.r-bloggers.com/2021/02/how-does-polychoric-correlation-work-aka-ordinal-to-ordinal-correlation/)
[An Alternative to the Correlation Coefficient That Works For Numeric and Categorical Variables](https://rviews.rstudio.com/2021/04/15/an-alternative-to-the-correlation-coefficient-that-works-for-numeric-and-categorical-variables/)
## Dummykodierung
[R Library Contrast Coding Systems for categorical variables](https://stats.oarc.ucla.edu/r/library/r-library-contrast-coding-systems-for-categorical-variables/)
[Dummykodierung](https://de.wikipedia.org/wiki/Dummy-Variable)
[fastDummies: Fast Creation of Dummy (Binary) Columns and Rows from Categorical Variables](https://cran.r-project.org/web/packages/fastDummies/)
## Analyse von Zeitreihen mit `{gamm4}`
Hier nochmal `{mgcv}` und `{gamm4}`
[Introduction to Generalized Additive Mixed Models](https://r.qcbs.ca/workshop08/book-en/introduction-to-generalized-additive-mixed-models-gamms.html)
`s()` und Interaktion mit `s(x_1, by = f_1)`
## Mediatoranalyse
[Causal Inference in R](https://www.r-causal.org/)
[Simulating confounders, colliders and mediators](https://freerangestats.info/blog/2023/06/04/causality-sims)
[Statistical Control Requires Causal Justification](https://journals.sagepub.com/doi/10.1177/25152459221095823)
[Causal influence and DAGs](https://dtkaplan.github.io/Lessons-in-statistical-thinking/L24-Causality-and-DAGS.html)
[Mediators, confounders, colliders – a crash course in causal inference](https://theoreticalecology.wordpress.com/2019/04/14/mediators-confounders-colliders-a-crash-course-in-causal-inference/)
[Collider Bias in Beobachtungsstudien: Konsequenzen für die medizinische Forschung](https://www.aerzteblatt.de/archiv/collider-bias-in-beobachtungsstudien-konsequenzen-fuer-die-medizinische-forschung-675e398b-1715-427e-b497-f210a57b605f)
[Thinking Clearly About Correlations and Causation: Graphical Causal Models for Observational Data](https://osf.io/preprints/psyarxiv/t3qub_v1)
[DAGitty — draw and analyze causal diagrams](https://www.dagitty.net/)
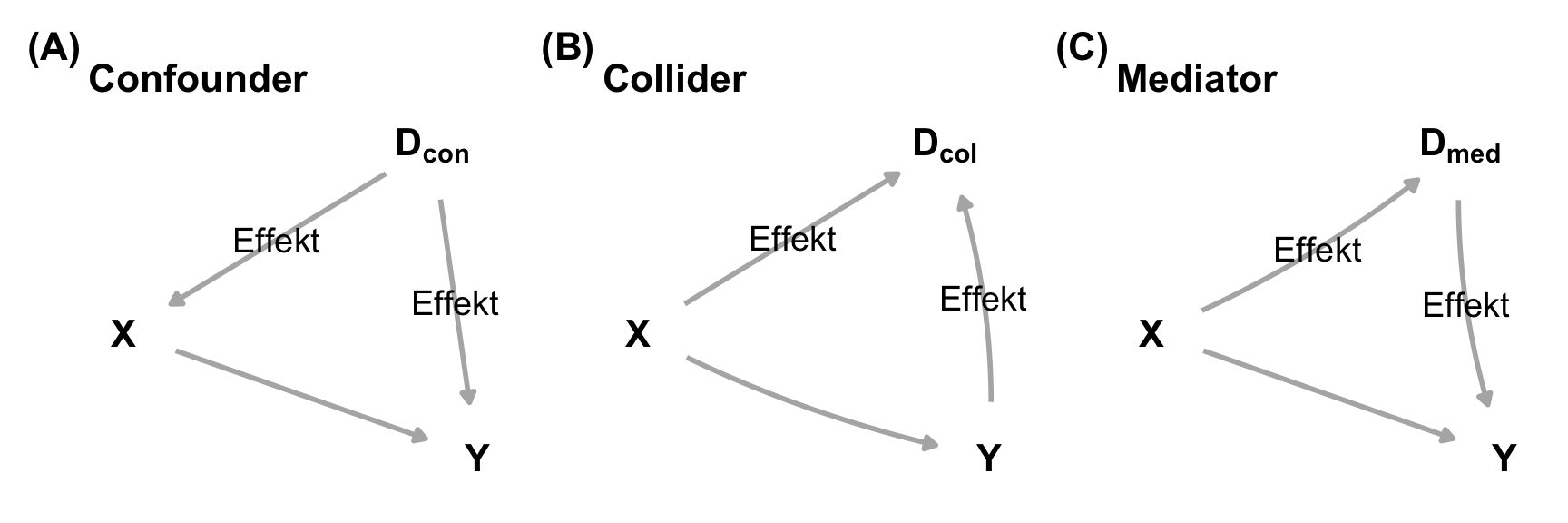
In dem Begriff des Disturber (deu. *Störenfried*, abk. $D$) fassen wir die Begriffe des Confounder (deu. *Störfaktor*, abk. $D_{con}$), Collider (deu. *Zusammenstoßen*, abk. $D_{col}$) und Mediator (deu. *Vermittlern*, abk. $D_{med}$) zusammen.
```{r}
#| message: false
#| echo: false
#| warning: false
#| fig-align: center
#| fig-height: 3
#| fig-width: 9
#| fig-cap: "foo **(A)** Confounder **(B)** Collider **(C)** Mediator. *[Zum Vergrößern anklicken]*"
#| label: fig-model-m-dag
arrow_col <- "grey70"
set.seed(123)
p1_dag <- dagify(
Y ~ X + C,
X ~ C
) %>%
ggplot(aes(x = x, y = y, xend = xend, yend = yend)) +
geom_dag_edges_fan(aes(label = c("Effekt", "Effekt", "", "")),
edge_colour = arrow_col, label_size = 5, edge_width = 1,
label_colour = "black") +
geom_dag_text(colour = "black", size = 5.5, parse = TRUE,
label = c(expression(bold(D[con])), expression(bold(X)), expression(bold(Y)))) +
theme_dag() +
labs(title = "Confounder") +
theme(plot.title = element_text(size = 16, face = "bold"),
plot.caption = element_text(face = "italic"))
set.seed(123)
p2_dag <-
dagify(
C ~ X + Y,
Y ~ X
) %>%
ggplot(aes(x = x, y = y, xend = xend, yend = yend)) +
geom_dag_edges_fan(aes(label = c("", "Effekt", "", "Effekt")),
edge_colour = arrow_col, label_size = 5, edge_width = 1,
label_colour = "black") +
geom_dag_text(colour = "black", size = 5.5, parse = TRUE,
label = c(expression(bold(D[col])), expression(bold(X)), expression(bold(Y)))) +
theme_dag() +
labs(title = "Collider") +
theme(plot.title = element_text(size = 16, face = "bold"),
plot.caption = element_text(face = "italic"))
set.seed(123)
p3_dag <-
dagify(
C ~ X,
Y ~ X + C
) %>%
ggplot(aes(x = x, y = y, xend = xend, yend = yend)) +
geom_dag_edges_fan(aes(label = c("Effekt", "Effekt", "", "")),
edge_colour = arrow_col, label_size = 5, edge_width = 1,
label_colour = "black") +
geom_dag_text(colour = "black", size = 5.5, parse = TRUE,
label = c(expression(bold(D[med])), expression(bold(X)), expression(bold(Y)))) +
theme_dag() +
labs(title = "Mediator") +
theme(plot.title = element_text(size = 16, face = "bold"),
plot.caption = element_text(face = "italic"))
p1_dag + p2_dag + p3_dag +
plot_layout(ncol = 3) +
plot_annotation(tag_levels = 'A', tag_prefix = '(', tag_suffix = ')') &
theme(plot.tag = element_text(size = 16, face = "bold"))
```
### Structural Equation Modeling {.unnumbered}
Passt das hier rein? Müssen wir nochmal überlegen.
@van2023best [tidySEM](https://cjvanlissa.github.io/tidySEM/index.html)
[Structural Equation Modeling](https://bookdown.org/bean_jerry/using_r_for_social_work_research/structural-equation-modeling.html)
[Introduction to structural equation modeling (sem) in r with lavaan](https://stats.oarc.ucla.edu/r/seminars/rsem/)
[Intro to structural equation modeling](https://rpubs.com/Agrele/SEM)
Schöne Diagramme [Structural Equation Models](https://advstats.psychstat.org/book/sem/index.php)
## Bayes Ideen {.unnumbered}
https://paul-buerkner.github.io/brms/articles/index.html
https://paul-buerkner.github.io/brms/articles/brms_nonlinear.html
https://bookdown.org/content/4857/
https://bayesat.github.io/lund2018/slides/andrey_anikin_slides.pdf
http://mjskay.github.io/tidybayes/articles/tidybayes.html
[Stan](https://mc-stan.org/)
[R Paket rstanarm](https://mc-stan.org/rstanarm/articles/index.html)
[R Paket tidyposterior](https://tidyposterior.tidymodels.org/)
[Half a dozen frequentist and Bayesian ways to measure the difference in means in two groups](https://www.andrewheiss.com/blog/2019/01/29/diff-means-half-dozen-ways/#bayesian-regression)
https://bookdown.org/marklhc/notes_bookdown/group-comparisons.html